Resumen
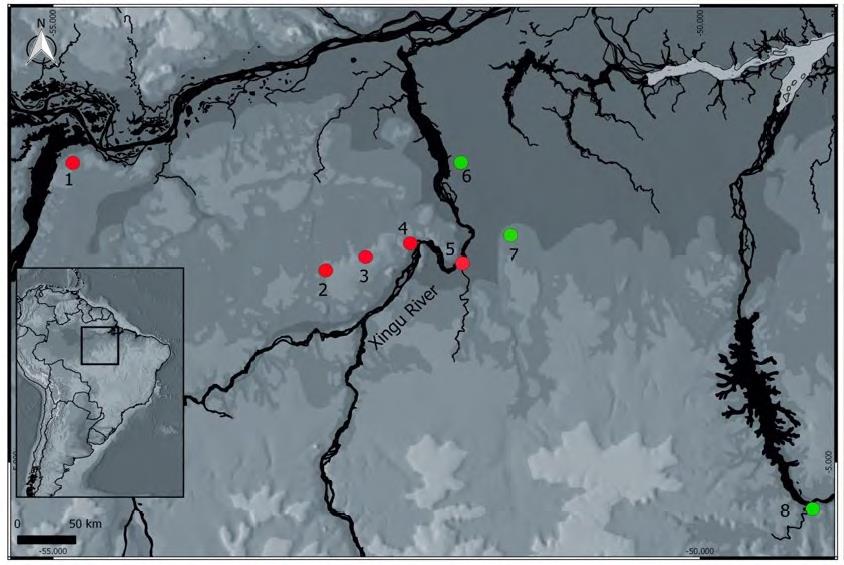
Una de las principales hipótesis para explicar el origen de la diversidad amazónica invoca el efecto de barrera de los ríos para explicar los patrones de diversidad. Esa hipótesis propone que algunos ríos pueden separar poblaciones continuas conduciendo a la diferenciación y la especiación. En ese sentido nos propusimos estudiar la estructura genética de Pristimantis latro, una especie recién descrita que habita una región bajo fuerte presión antrópica producto de la ganadería expansiva, la minería ilegal y la construcción de represas hidroeléctricas. El ADN se extrajo de 52 individuos de P. latro y se amplificó mediante reacción en cadena de la polimerasa (PCR) usando los marcadores mitocondriales 16S rRNA y COI. Para inferir el tiempo de divergencia entre las localidades de P. latro, construimos un árbol de especies e hicimos análisis de varianza molecular (AMOVA) para inferir la diferenciación genética entre y dentro de las poblaciones de P. latro. Encontramos que P. latro presenta una estructuración genética en las poblaciones de las márgenes derecha e izquierda del río Xingu y dentro de las regiones interfluviales deTapajós-Xingu y Xingu-Tocantins. Asimismo, el tiempo de divergencia entre las poblaciones de las márgenes derecha e izquierda del río Xingu aconteció hace aproximadamente 380.000 años. El patrón de estructura genética que se encontró se corresponde con el indicado en artículos recientes para el género Pristimantis, el cual revela que las especies sin renacuajos exhiben una estructura genética que responde a la hipótesis de los ríos como barreras.
Referencias
Almeida, A.S. de, Vieira, I.C.G., Barros, M.N.R., Rocha, D.P.N. (2014). Área de endemismo Belém e Xingu: configuração e espacialização do uso da terra e da cobertura vegetal. In: Thaise Emilio; Flávio Luizão. (Org.). Cenários para Amazônia: clima, biodiversidade e uso da terra. 1ed.Manaus: INPA, p. 57-66.
Andersen, L.W., Fog, K., Damgaard, C. (2004). Habitat fragmentation causes bottlenecks and inbreeding in the European tree frog (Hyla arborea). Proceedings of the Royal Society B: Biological Sciences. 271: 1293-1302. Doi: 10.1098/rspb.2004.2720
Antonelli, A., Quijada-Mascareñas, A., Crawford, A.J., Bates, J.M., Velazco, P.M., Wuster, W. (2010). Molecular studies and phylogeography of Amazonian tetrapods and their relation to geological and climatic models. Molecular studies of Amazonian tetrapods. En C. Hoorn and F. P. Wesselingh (Ed), Amazonia: Landscape and Species Evolution: A look into the past (pp. 385-404). Wiley-Blackwell
Bates, H. (1874). The naturalist on the River Amazon. John Murray, London. 506 pp.
Beebee, T.J.C., Griffiths, R.A. (2005). The amphibian decline crisis: A watershed for conservation biology? Biological Conservation. 125: 271-285. Doi: 10.1016/j.biocon.2005.04.009
Bogart, J. P. (1991). The influence of life history on karyotypic evolution in frogs. Amphibian Cytogenetics and Evolution. Academic Press, San Diego. p. 233-258.
Burns, E.L., Eldridge, M.D.B., Houlden, B.A. (2004). Microsatellite variation and population structure in a declining Australian Hylid Litoria aurea. Molecular Ecology. 13: 1745-1757. Doi: 10.1111/j.1365-294X.2004.02190.x
Corander, J., Marttinen, P., Sirén, J., Tang, J. (2008). Enhanced Bayesian modelling in BAPS software for learning genetic structures of populations. BMC bioinformatics. 9: 1-14. Doi: 10.1186/1471-2105-9-539
Da Rocha, D.G., Kaefer, I.L. (2019). What has become of the refugia hypothesis to explain biological diversity in Amazonia? Ecology and Evolution. 9: 4302-4309. Doi:10.1002/ece3.5051
Darriba, D., Taboada, G.L., Doallo, R., Posada, D. (2012). JModelTest 2: More models, new heuristics and parallel computing. Nature Methods. 9: 772. Doi: 10.1038/nmeth.2109
Doyle, J.J., Doyle, J.L. (1987). Isolation of plant DNA from fresh tissue. Focus. 12 (1): 13-15.
Drummond, A.J., Ho, S.Y.W., Phillips, M.J., Rambaut, A. (2006). Relaxed phylogenetics and dating with confidence. PLoS Biology. 4: 699-710. Doi: 10.1371/journal.pbio.0040088
Drummond, A.J., Rambaut, A. (2007). BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 7: 1-8. Doi: 10.1186/1471-2148-7-214
Drummond, A, J., Suchard, M.A., Xie, D., Rambaut, A. (2012). Bayesian Phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 29: 1969-1973. Doi: 10.1093/molbev/mss075
Excoffier, L., Laval, G., Schneider, S. (2005). Arlequin 3.0: An integrated software package for population genetics data analysis. Evolution Bioinformatics Online. 1: 47-50.
Felsenstein, J. (1985). Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution. 39: 783-791. Doi: 10.2307/2408678
Folmer, O., Black, M., Hoeh, W., Lutz, R., Vrijenhoek, R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology Biotechnology. 3: 294-299.
Fouquet, A., Ledoux, J.B., Dubut, V., Noonan, B.P., Scotti, I. (2012). The interplay of dispersal limitation, rivers, and historical events shapes the genetic structure of an Amazonian frog. Biological Journal of the Linnean Society. 106: 356-373. Doi: 10.1111/j.1095-8312.2012.01871.x
Fouquet, A., Martínez, Q., Courtois, E.A., Dewynter, M., Pineau, K., Gaucher, P., Blanc, M., Marty, C., Kok, P.J.R. (2013). A new species of the genus Pristimantis (Amphibia, Craugastoridae) associated with the moderately evelated massifs of French Guiana. Zootaxa. 3750: 569-586. Doi: 10.11646/zootaxa.3750.5.8
Fouquet, A., Courtois, E.A., Baudain, D., Lima, J.D., Souza, S.M., Noonan, B.P., Rodrigues, M.T. (2015). The trans-riverine genetic structure of 28 Amazonian frog species is dependent on life history. Journal of Tropical Ecology. 31: 361-373. Doi: 10.1017/S0266467415000206
Fourdrilis, S., Mardulyn, P., Hardy, O.J., Jordaens, K., Freitas Martins, A.M. de, Backeljau, T. (2016). Mitochondrial DNA hyperdiversity and its potential causes in the marine periwinkle Melarhaphe neritoides (Mollusca: Gastropoda). PeerJ. 4: e2549. Doi: 10.7717/peerj.2549
Funk, W.C., Caldwell, J.P., Peden, C.E., Padial, J.M., De la Riva, I., Cannatella, D.C. (2007). Tests of biogeographic hypotheses for diversification in the Amazonian forest frog, Physalaemus petersi. Molecular Phylogenetics and Evolution. 44: 825-837. Doi: 10.1016/j. ympev.2007.01.012
Funk, W.C., Caminer, M., Ron, S.R. (2012). High levels of cryptic species diversity uncovered in Amazonian frogs. Proceedings of the Royal Society of London. Series B, Biological sciences. 279: 1806-1814. Doi: 10.1098/rspb.2011.1653
Guayasamin, J.M., Hutter, C.R., Tapia, E.E., Culebras, J., Pyron, R.A., Morochz, C., Funk, W.C., Arteaga, A. (2017). Diversification of the rainfrog Pristimantis ornatissimus in the lowlands and Andean foothills of Ecuador. PLoS ONE. 12: 1-21.
Hall, T.A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium: 95-98.
Heinicke, M.P., Duellman, W.E., Trueb, L., Means, D.B., Macculloch, R.D., Hedges, S.B. (2009). A new frog family (Anura: Terrarana) from South America and an expanded directdeveloping clade revealed by molecular phylogeny. Zootaxa. 2111: 1-35. Doi: 10.11646/zootaxa.2211.1.1
Instituto Brasileiro de Geografia e Estatistica - IBGE (2012). Manual técnico da vegetação brasileira. Rio de Janeiro. 271p.
Jobb, G., Haeseler, A. V., Strimmer, K. (2011). TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evolutionary Biology, 4:18. doi: 10.1186/1471-2148-4-18.
Kaefer, I.L., Kaefer, I.L., Tsuji-Nishikido, B.M., Lima, A.P. (2012). Beyond the river: Underlying determinants of population acoustic signal variability in Amazonian direct-developing Allobates (Anura: Dendrobatoidea). Acta Ethologica. 15: 187-194. Doi: 10.1007/s10211-012-0126-0
Kaefer, I.L., Tsuji-Nishikido, B.M., Mota, E.P., Farias, I.P., Lima, A.P. (2013). The Early Stages of Speciation in Amazonian Forest Frogs: Phenotypic Conservatism despite Strong Genetic Structure. Evolutionary Biology. 40: 228-245. Doi: 10.1007/s11692-012-9205-4.
Lampert, K.P., Rand, A.S., Mueller, U.G., Ryan, M.J. (2003). Fine-scale genetic pattern and evidence for sex-biased dispersal in the túngara frog, Physalaemus pustulosus. Molecular Ecology. 12: 3325-3334. Doi: 10.1046/j.1365-294X.2003.02016.x
Lemmon, E.M., Lemmon, A.R., Collins, J.T., Cannatella, D.C. (2008). A new North American chorus frog species (Amphibia: Hylidae: Pseudacris) from the south-central United States. Zootaxa. 1675: 1-30.
Lymberakis, P., Poulakakis, N., Manthalou, G., Tsigenopoulos, C.S., Magoulas, A., Mylonas, M. (2007). Mitochondrial phylogeography of Rana (Pelophylax) populations in the Eastern Mediterranean region. Molecular Phylogenetics and Evolution. 44: 115-125. Doi: 10.1016/j. ympev.2007.03.009
Palumbi, S.R., Martin, A., Romano, S., McMillan, W.O., Stice, L., Grabowski, G. (1991) ‘‘The Simple Fool’s Guide to PCR, Version 2.0.’’ Privately published document compiled by S. Palumbi, Dept. Zoology, Univ. Hawaii.
Miller, M.P. (2005). Alleles In Space (AIS): Computer Software for the Joint Analysis of Interindividual Spatial and Genetic Information. Journal of Heredity. 96: 722-724. Doi: 10.1093/jhered/esi119
Moraes, L.J.C.L., Pavan, D., Barros, M.C., Ribas, C.C. (2016). The combined influence of riverine barriers and flooding gradients on biogeographical patterns for amphibians and squamates in south-eastern Amazonia. Journal of Biogeography. 43: 2113-2124. Doi: 10.1111/jbi.12756
Oliveira, E.A., Rodrigues, L.R., Kaefer, I.L., Pinto, K.C., Hernández-Ruz, E.J. (2017). A new species of Pristimantis from eastern Brazilian Amazonia (Anura: Craugastoridae). ZooKeys. 2017: 101-129. Doi: 10.3897/zookeys.687.13221
Oliveira, E.A., Silva, L.A., Silva, E.A.P., Guimaraes, K.L.A., Penhacek, M.P., Martínez, J.G., Rodrigues, L.R.R., Santana, D.J., Hernández-Ruz, E.J. (2020). Four new species of Pristimantis Jiménez de la Espada, 1870 (Anura: Craugastoridae) in the eastern Amazon. PLoS One. 15 (3): e0229971.
Ribas, C.C., Aleixo, A., Nogueira, A.C.R., Miyaki, C.Y., Cracraft, J., Andre, A. (2012). A palaeobiogeographic model for biotic diversification within Amazonia over the past three million years. Proceedings of the Royal Society of London. Series B, Biological sciences. 279: 681-689. Doi: 10.1098/rspb.2011.1120
Rodríguez, A., Bomer, M., Pabijan, M., Gehara, M., Haddad, C.F.B. (2015). Genetic divergence in tropical anurans: Deeper phylogeographic structure in forest specialists and in topographically complex regions. Evolutionary Biology. 29: 765-785.
Salzburger, W., Ewing, G.B., von Haeseler, A. (2011). The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Molecular Ecology. 20: 1952-1963. Doi: 10.1111/j.1365-294X.2011.05066.x
Simões, P.I., Stow, A., Hödl, W., Amézquita, A., Farias, I.P., Lima, A.P. (2014). The value of including intraspecific measures of biodiversity in environmental impact surveys is highlighted by the Amazonian brilliant-thighed frog (Allobates femoralis). Tropical Conservation Science. 7: 811-828. Doi: 10.1177/194008291400700416
Tamura, K., Stecher, G., Peterson, D., Filipski, A., Kumar, S. (2013). MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 30: 2725-2729. Doi: 10.1093/molbev/mst197
Thompson, J.D., Higgins, D.G., Gibson, T.J. (1996). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 22: 4673-4680. Doi: 10.1093/nar/22.22.4673
Vieites, D.R., Wollenberg, K.C., Andreone, F., Kohler, J., Glaw, F., Vences, M. (2009) Vast underestimation of Madagascar’s biodiversity evidenced by an integrative amphibian inventory. Proceedings of the National Academy of Sciences. 106: 8267-8272. Doi: 10.1073/pnas.0810821106
Vos, C.C., Antonisse-de Jong, A.G., Goedhart, P.W., Smulders, M.J.M. (2001). Genetic similarity as a measure for connectivity between fragmented populations of the moor frog (Rana arvalis). Heredity. 86: 598 - 608. PMID: 11554976
Wallace, A.R. (1852). On the Monkeys of the Amazon. Zoological Society of London. 20: 107-110.
Wells, K.D. (2007). The Ecology and Behavior of Amphibians. Chicago, USA: The University of Chicago Press. Pp. 1148
Wright, S. (1943). Isolation by distance. Genetics. 28: 114-138. Doi: 10.5194/isprs-Archives-XLII-5-W1-419-2017

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-SinDerivadas 4.0.
Derechos de autor 2020 Revista de la Academia Colombiana de Ciencias Exactas, Físicas y Naturales