Resumen
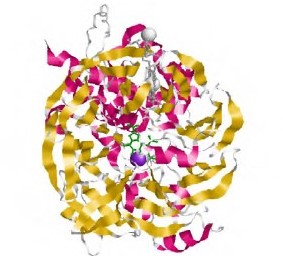
Entre las aplicaciones del modelamiento por homología están la predicción de ligandos, el análisis de mutaciones, y las actividades catalíticas. Normalmente, el uso de varios moldes produce modelos más precisos en la medida en que capturan mejor las variantes estructurales entre todas las posibles conformaciones de las proteínas de una familia. El modelamiento comparativo es una estrategia útil cuando no se tiene información en las ases de datos y se quiere hacer un ensayo xperimental relacionado con la estructura y el funcionamiento de una proteína. En el presente trabajo se analizó estructural y funcionalmente la quinoproteína glucosa deshidrogenasa unida a la membrana (membrane-bound PQQ-dependent glucose dehydrogenase, mGDH, PQQ mGDH) en Pseudomonas fluorescens, reconocida como un microorganismo promotor del crecimiento vegetal y una de cuyas funciones es la solubilización de fosfatos inorgánicos catalizados mediante PQQ mGDH. Esta es la primera enzima que participa en la oxidación directa de la glucosa transformando la D-lucosa a D-gluconato, proceso que es el inicio en la producción de ácidos orgánicos implicados en la solubilización de fosfatos inorgánicos. A pesar de la gran cantidad de estudios con esta enzima, no se ha reportado la determinación de su estructura tridimensional de la enzima quinoproteína glucosa deshidrogenasa (PQQ-GDH) en la bacteria P. fluorescens. En este estudio se comparó la secuencia e la proteína con las reportadas en las bases de datos para otras especies del género Pseudomonas y se propuso un modelo tridimensional de la enzima PQQ mGDH en P. fluorescens mediante el modelamiento por homología para caracterizar su estructura secundaria, el dominio catalítico, y el sitio de unión al ofactor pirroloquinolina quinona (PQQ), así como los dominios funcionales presentes en la proteína.
Referencias
Alori, E. T., Glick, B. R., & Babalola, O. O. (2017). Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Frontiers in Microbiology. 8: 971. Doi: 10.3389/fmicb.2017.00971
Ameyama, M., Matsushita, K., Shinagawa, E., & Adachi, O. (1991). Biochemical and physiological functions of pyrroloquinoline quinone. Vitamins and Hormones. 46: 229-270. http://www.ncbi.nlm.nih.gov/pubmed/1660640
An, R., & Moe, L. A. (2016). Regulation of Pyrroloquinoline Quinone-Dependent Glucose Dehydrogenase Activity in the Model Rhizosphere-Dwelling Bacterium Pseudomonas putida KT2440.
Applied and Environmental Microbiology. 82 (16): 4955-4964. Doi: 10.1128/aem.00813-16 Anthony, C. (2001). Pyrroloquinoline quinone (PQQ) and quinoprotein enzymes. Antioxidants and Redox Signaling. 3 (5): 757-774. Doi: 10.1089/15230860152664966
Anthony, C., & Zatman, L. (1967a). The microbial oxidation of methanol. The prosthetic group of the alcohol dehydrogenase of Pseudomonas sp. M27: a new oxidoreductase prosthetic group. Biochemical Journal. 104 (3): 960-969. Doi: 10.1042/bj1040960
Anthony, C., & Zatman, L. J. (1967b). The microbial oxidation of methanol. The prosthetic group of the alcohol dehydrogenase of Pseudomonas sp. M27: a new oxidoreductase prosthetic group. The Biochemical Journal. 104 (3): 960-969. Doi: 10.1042/bj1040960
Asahi, Y., Noiri, Y., Igarashi, J., Asai, H., Suga, H., Ebisu, S. (2010). Effects of N-acyl homoserine lactone analogues on Porphyromonas gingivalis biofilm formation. Journal of Periodontal Research. 45 (2): 255-261. Doi: 10.1111/j.1600-0765.2009.01228.x
Babnigg, G., & Joachimiak, A. (2010). Predicting protein crystallization propensity from protein sequence. Journal of Structural and Functional Genomics. 11 (1): 71-80. Doi: 10.1007/s10969-010-9080-0
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., Noble, W. S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research. 37:W202-W208. Doi: 10.1093/nar/gkp335
Ben Farhat, M., Fourati, A., Chouayekh, H. (2013). Coexpression of the pyrroloquinoline quinone and glucose dehydrogenase genes from Serratia marcescens CTM 50650 conferred high mineral phosphate-solubilizing ability to Escherichia coli. Applied Biochemistry and Biotechnology. 170 (7): 1738-1750. Doi: 10.1007/s12010-013-0305-0
Benkert, P., Künzli, M., Schwede, T. (2009). QMEAN server for protein model quality estimation. Nucleic Acids Research: 37 (suppl_2): W510-W514. Doi: 10.1093/nar/gkp322
Chapman, H. N., Fromme, P., Barty, A., White, T. a, Kirian, R. a, Aquila, A., Spence, J. C. H. (2011). Femtosecond X-ray protein nanocrystallography - Supplementary Information. Nature. 470 (7332): 73-77. Doi: 10.1038/nature09750
Chen, C. K.-M., Chan, N.-L., Wang, A. H.-J. (2011). The many blades of the β-propeller proteins: conserved but versatile. Trends in Biochemical Sciences. 36 (10): 553-561. Doi: 10.1016/j.tibs.2011.07.004
Combet, C., Jambon, M., Deleage, G., Geourjon, C. (2002). Geno3D: automatic comparative molecular modelling of protein. Bioinformatics. 18 (1): 213-214. Doi: 10.1093/bioinformatics/18.1.213
Cozier, G. E., Salleh, R. A., Anthony, C. (1999). Characterization of the membrane quinoprotein glucose dehydrogenase from Escherichia coli and characterization of a site-directed mutant in which histidine-262 has been changed to tyrosine. The Biochemical Journal. 340 (Pt 3): 639-647. http://www.ncbi.nlm.nih.gov/pubmed/10359647
Dassault Systèmes BIOVIA. (2019). Discovery studio. San Diego: Dessault Systemes.
Duine, J. A. (1991). Quinoproteins: enzymes containing the quinonoid cofactor pyrroloquinoline quinone, topaquinone or tryptophan‐tryptophan quinone. European Journal of Biochemistry. 200 (2): 271-284. Doi: 10.1111/j.1432-1033.1991.tb16183.x
Duine, J. A. & Jongejan, J. A. (1989). Quinoproteins, enzymes with pyrrolo-quinoline quinone as cofactor. Annual Review of Biochemistry. 58: 403-426. Doi: 10.1146/annurev.bi.58.070189.002155
Goldstein, A., Lester, T., Brown, J. (2003). Research on the metabolic engineering of the direct oxidation pathway for extraction of phosphate from ore has generated preliminary evidence for PQQ biosynthesis in Escherichia coli as well as a possible role for the highly conserved region of quinoprote. Biochimica et Biophysica Acta - Proteins and Proteomics. 1647 (1-2): 266-271. Doi: 10.1016/S1570-9639(03)00067-0
Good, N. M., Vu, H. N., Suriano, C. J., Subuyuj, G. A., Skovran, E., Martínez-Gómez, N. C. (2016). Pyrroloquinoline quinone ethanol dehydrogenase in Methylobacterium extorquens AM1 extends lanthanide-dependent metabolism to multicarbon substrates. Journal of Bacteriology. 198 (22): 3109-3118. Doi: 10.1128/JB.00478-16
Hamilton, N. & Huber, T. (2008). An introduction to protein contact prediction. Methods in Molecular Biology (Clifton, N.J.). 453: 87-104. Doi: 10.1007/978-1-60327-429-6_3
Hauge, J. G. (1964). Glucose Dehydrogenase of Bacterium with a Novel Prosthetic Group * an Enzyme. The Journal of Biological Chemistry. 239 (11): 3630-3639. http://www.jbc.org/
Hiraoka, T., Clark, J. I., LI, X. Y., Thurston, G. M. (1996). Effect of selected anti-cataract agents on opacification in the selenite cataract model. Experimental Eye Research: 62 (1): 11-19.Doi: 10.1006/exer.1996.0002
Itoh, S, Kawakami, H., Fukuzumi, S. (1998). Model studies on calcium-containing quinoprotein alcohol dehydrogenases. Catalytic role of Ca2+ for the oxidation of alcohols by coenzyme PQQ (4,5-dihydro-4,5-dioxo-1H-pyrrolo[2,3-f]quinoline-2, 7,9-tricarboxylic acid). Biochemistry. 37 (18): 6562-6571. Doi: 10.1021/bi9800092
Itoh, Shinobu, Kawakami, H., Fukuzumi, S. (2000). Development of the active site model for calcium-containing quinoprotein alcohol dehydrogenases. Journal of Molecular Catalysis - BEnzymatic. 8 (1-3): 85-94. Doi: 10.1016/S1381-1177(99)00070-3
Kim, H.-W., Wang, J.-Y., Lee, J.-Y., Park, A.-K., Park, H., Jeon, S.-J. (2016). Biochemical and structural characterization of quinoprotein aldose sugar dehydrogenase from Thermus thermophilus HJ6: Mutational analysis of Tyr156 in the substrate-binding site. Archives of Biochemistry and Biophysics. 608: 20-26. Doi: 10.1016/j.abb.2016.08.022
Krishnaraj, P. U. & Goldstein, A. (2002). Cloning of a Serratia marcescens DNA fragment that induces quinoprotein glucose dehydrogenase-mediated gluconic acid production in Escherichia coli in the presence of stationary phase Serratia marcescens. FEMS Microbiology Letters. 205 (2): 215-220. Doi: 10.1016/s0378-1097(01)00472-4
Kuriyan, J., Konforti, B., Wemmer, D. (2013). The molecules of life: physical and chemical principles. Garland Science. 1032 p. New York.
Laurinavicius, V., Kurtinaitiene, B., Stankeviciute, R. (2008). Behavior of PQQ glucose dehydrogenase on Prussian blue-modified carbon electrode. Electroanalysis. 20 (13): 1391-1395. Doi: 10.1002/elan.200804216
Lidbury, I. D. E. A., Murphy, A. R. J., Scanlan, D. J., Bending, G. D., Jones, A. M. E., Moore, J. D., Wellington, E. M. H. (2016). Comparative genomic, proteomic and exoproteomic analyses of three Pseudomonas strains reveals novel insights into the phosphorus scavenging capabilities of soil bacteria. Environmental Microbiology. 18 (10): 3535-3549. Doi: 10.1111/1462-2920.13390
Lüthy, R., Bowie, J. U., Eisenberg, D. (1992). Assessment of protein models with three-dimensional profiles. Nature. 356 (6364): 83-85. Doi: 10.1038/356083a0
Madeira, F., Park, Y. mi, Lee, J., Buso, N., Gur, T., Madhusoodanan, N., López, R. (2019). The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Research. 47(W1): W636-W641. Doi: 10.1093/nar/gkz268
McGuffin, L. J., Bryson, K., Jones, D. T. (2000). The PSIPRED protein structure prediction server. Bioinformatics. 16 (4): 404-405. Doi: 10.1093/bioinformatics/16.4.404
McIntire, W. S. (1998). Newly discovered redox cofactors: possible nutritional, medical, and pharmacological relevance to higher animals. Annual Review of Nutrition. 18: 145-177. Doi: 10.1146/annurev.nutr.18.1.145
Nishigori, H., Yasunaga, M., Mizumura, M., Lee, J. W., Iwatsuru, M. (1989). Preventive effects of pyrroloquinoline quinone on formation of cataract and decline of lenticular and hepatic glutathione of developing chick embryo after glucocorticoid treatment. Life Sciences. 45 (7):593-598. Doi: 10.1016/0024-3205(89)90044-1
Pérez, S., Coto, O., Echemendía, M., Ávila, G. (2015). Pseudomonas fluorescens Migula, ¿control biológico o patógeno? Rev. Protección Veg. 30 (3): 225-234.
Pons, T., Gómez, R., Chinea, G., Valencia, A. (2012). Beta-propellers: Associated Functions and their Role in Human Diseases. Current Medicinal Chemistry. 10 (6): 505-524. Doi:10.2174/0929867033368204
Ramachandran, G. N., Ramakrishnan, C., Sasisekharan, V. (1963). Stereochemistry of polypeptide chain configurations. Journal of Molecular Biology. 7: 95-99. Doi: 10.1016/S0022-2836(63)80023-6
Sakamoto, H., Uchii, T., Yamaguchi, K., Koto, A., Takamura, E.-I., Satomura, T., … Suye, S.-I. (2015). Construction of a biocathode using the multicopper oxidase from the hyperthermophilic archaeon, Pyrobaculum aerophilum: towards a long-life biobattery. Biotechnology Letters. 37 (7): 1399-1404. Doi: 10.1007/s10529-015-1819-z
Salisbury, S. A., Forrest, H. S., Cruse, W. B. T., Kennard, O. (1979). A novel coenzyme from bacterial primary alcohol dehydrogenases [13]. Nature. 280: 843-844. Doi: 10.1038/280843a0
Sánchez-Garcés, M. A., Álvarez-Camino, J. C., Corral-Pavón, E., González-Martínez, R., Alves-Marques, J., Párraga-Manzol, G., Gay-Escoda, C. (2012). Revisión bibliográfica de Implantología Bucofacial del año 2010: Segunda parte. Avances En Periodoncia e Implantología Oral. 24 (2): 77-94. Doi: 10.4321/s1699-65852012000200003
Sayle, R. (1995). RASMOL: biomolecular graphics for all. Trends in Biochemical Sciences. 20 (9): 374-376. Doi: 10.1016/S0968-0004(00)89080-5
Sippl, M. J. (1993). Recognition of errors in three-dimensional structures of proteins. Proteins: Structure, Function, and Genetics. 17 (4): 355-362. Doi: 10.1002/prot.340170404
Takeda, K., Ishida, T., Yoshida, M., Samejima, M., Ohno, H., Igarashi, K., Nakamura, N. (2019). Crystal Structure of the Catalytic and Cytochrome b Domains in a Eukaryotic Pyrroloquinoline Quinone-Dependent Dehydrogenase. Applied and Environmental Microbiology. 85 (24): 1-13. Doi: 10.1128/AEM.01692-19
Vera-Cardoso, B. C., Muñoz-Rojas, J., Munive, J. A., Marín-Cevada, V., Flores-Encarnación, M., & Ricardo, C. (2017). Pirroloquinolinaquinona (PQQ) y las bacterias promotoras del crecimiento vegetal (PGPR). De la biosíntesis a los fenotipos. Alianzas & Tendencias.2 (1): 22-29.
Wen, Z. T., Nguyen, A. H., Bitoun, J. P., Abranches, J., Baker, H. V, & Burne, R. A. (2011). Transcriptome analysis of LuxS-deficient Streptococcus mutans grown in biofilms. Molecular Oral Microbiology. 26 (1): 2-18. Doi: 10.1111/j.2041-1014.2010.00581.x
Wiederstein, M., & Sippl, M. J. (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research. 35: 407-410. Doi:10.1093/nar/gkm290
Yamamoto, M., Hirata, K., Hikima, T., Kawano, Y., Ueno, G. (2010). Protein micro-crystallography with a new micro-beam beamline. Yakugaku Zasshi : Journal of the Pharmaceutical Society of Japan. 130 (5): 641-648. Doi: 10.1248/yakushi.130.641
Zdobnov, E. M. & Apweiler, R. (2001). InterProScan - An integration platform for the signaturerecognition methods in InterPro. Bioinformatics. 17 (9): 847-848. Doi: 10.1093/bioinformatics/17.9.847

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-SinDerivadas 4.0.
Derechos de autor 2020 Revista de la Academia Colombiana de Ciencias Exactas, Físicas y Naturales