Resumen
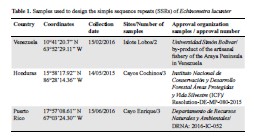
Como primer paso para establecer la estructura genética del erizo de mar Echinometra lucunter lucunter a lo largo del Caribe, se aislaron 26 microsatélites usando secuenciación de extremo pareado (Next Generation Sequencing, NGS) Illumina. Se optimizaron exitosamente 17 marcadores y se probó su variación alélica en 23 individuos recolectados a lo largo del mar Caribe y en Cabo Verde, Atlántico oriental tropical. El número de alelos por locus (Na) fluctuó entre cuatro y 24, la heterocigosidad observada (Ho) entre 0,682 y 1 y la heterocigosidad esperada (He), entre 0,609 y 0,9304. No hubo desequilibrio de ligamento entre los pares de locus detectados. Los microsatelites aislados e identificados se usarán por primera vez para detectar la influencia de las barreras marinas en el flujo génico del erizo E. lucunter lucunter a lo largo del mar Caribe. Estos nuevos marcadores serán esenciales para la conservación y los estudios de conectividad a través del mar Caribe y el océano Atlántico, área donde se distribuye la especie.
Referencias
Avise, J. C. (1992). Molecular population structure and biogeographic history of a regional fauna a case history with lessons for conservative biology. Oikos. 63: 62-76. Doi: 10.2307/3545516
Baums, I. B., Miller, M. W., Hellberg, M. E. (2005). Regionally isolated populations of an imperiled Caribbean coral, Acropora palmata. Mol Ecol. 14: 1377-1390. Doi: 10.1111/j.1365-294X.2005.02489.x
Blacket, M. J., Robin. C., Good, R. T, Lee, S. F, Miller, A. D. (2012). Universal primers for fluorescent labelling of PCR fragments – an effective approach to genotyping by fluorescence. Mol Ecol Resour. 1: 456-463. Doi: 10.1111/j.1755-0998.2011.03104.x
Carlin, J. L., Robertson, D. R., Bowen, B. W. (2003). Ancient divergences and recent connections in two tropical Atlantic reef fishes Epinephelus adscensionis and Rypticus saponaceous (Percoidei: Serranidae). Mar Biol. 143: 1057-1069. Doi: 10.1007/s00227-003-1151-3
Cowen, R. K., Paris, C. B., Srinivasan, A. (2006). Scaling of population connectivity in marine populations. Sci. 311: 522-527. Doi: 10.1126/science.1122039
Culley, T. M., Stamper, T. I., Stokes, R. L., Brzski, J. R., Hardman, N. A., Klooster, M. R, Merritt, B. J. (2013). An efficient technique for primer development and application that integrates fluorescent labelling and multiplex PCR. Appl Plant Sci. 1 (10): 1-10. Doi: 10.3732/apps.1300027
Fox, G., Preziosi, R. F., Antwis, R. E., Benavides‐Serrato M., Combe, F. J., Harris, W. E., Hartley, I. R., Kitchener, A. C., De Kort, S. R., Nekaris, A. I., Rowntree, J. K. (2019). Multi‐individual microsatellite identification: A multiple genome approach to microsatellite design (MiMi). Mol Ecol Resour. 19 (6): 1-9. Doi: 10.1111/1755-0998.13065
Griffiths, S. M., Fox, G., Briggs, P. J., Donaldson, I. J., Hood, S., Richardson, P., Leaver, G. W., N. K. Truelove, Preziosi, R. F. (2016). A Galaxy-based bioinformatics pipeline for optimised, streamlined microsatellite development from Illumina next-generation sequencing data. Conserv Genet Resour. 8: 481-486. Doi: 10.1007/s12686-016-0570-7
Kalinowski, S. T., Taper, M. L., Marshall, T.C. (2007). Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol. 16: 1099-1106. Doi: 10. 1111/j.1365-294X.2007.03089.x
McCartney, M. A., Keller, G., Lessios, H. A. (2000). Dispersal barriers in tropical oceans and speciation in Atlantic and eastern Pacific sea urchins of the genus Echinometra. Mol. Ecol. 9: 1391-1400. Doi: 10.1046/j.1365-294x.2000.01022.x
Paris, C. B., Cowen, R. K. (2004). Direct evidence of a biophysical retention mechanism for coral reef fish larvae. Limnol. 49 (6): 1964-1979. Doi: 10.4319/lo.2004.49.6.1964
Pawson, D. L. (1978). The echinoderm fauna of Ascension Island, South Atlantic Ocean. Smithsonian Contrib. Mar. Sci, 2: 1-29.
Qiagen. DNeasy® Blood & Tissue Handbook [pdf] (2006). Date of entry: January 15, 2015. Available in: http://mvz.berkeley.edu/egl/inserts/DNeasy_Blood_&_Tissue_Handbook.pdf
Qiagen. Type-it Microsatellite PCR Handbook [pdf] (2009). Date of entry: March 14, 2015. Available in: http://www.qiagen.com/Products/Catalog/Assay-Technologies/End-Point-PCR-and-RTPCR-Reagents/Type-it-Microsatellite-PCR-Kit#resources
Schultz, E. T. & Cowen, R. K. (1994). Recruitment of coral-reef fishes to Bermuda – local retention or long-distance transport. Mar Ecol Prog Ser. 109: 15-28. Doi: 10.3354/meps111015
Selkoe, K. A. & Toonen, R. J. (2006). Microsatellites for ecologists: A practical guide to using and evaluating microsatellite markers. Ecol Lett. 9: 615-29. Doi: 10.1111/j.1461-0248.2006.00889.x
Sunnucks, P. (2000). Efficient genetic markers for population biology. Trends Ecol Evol. 15 (5): 199-203. Doi: 10.1016/S0169-5347(00)01825-5
Taylor, M. S. & Hellberg, M. E. (2003). Genetic evidence for local retention of pelagic larvae in a Caribbean reef fish. Sci. 299: 107-109. Doi: 10.1126/science.1079365
Taylor, M. S. & Hellberg, M. E. (2006). Comparative phylogeography in a genus of coral reef fishes: biogeographic and genetic concordance in the Caribbean. Mol Ecol. 15: 695-707. Doi: 10.1111/j.1365-294X.2006.02820.x
Van Oosterhout, C., Hutchison, W. F., Shipley, P., Wills, D. P. M. (2004). Micro-Checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Notes. 4: 535-538. Doi: 10.1111/j.1471-8286.2004.00684.x

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-SinDerivadas 4.0.
Derechos de autor 2020 Revista de la Academia Colombiana de Ciencias Exactas, Físicas y Naturales