Resumen
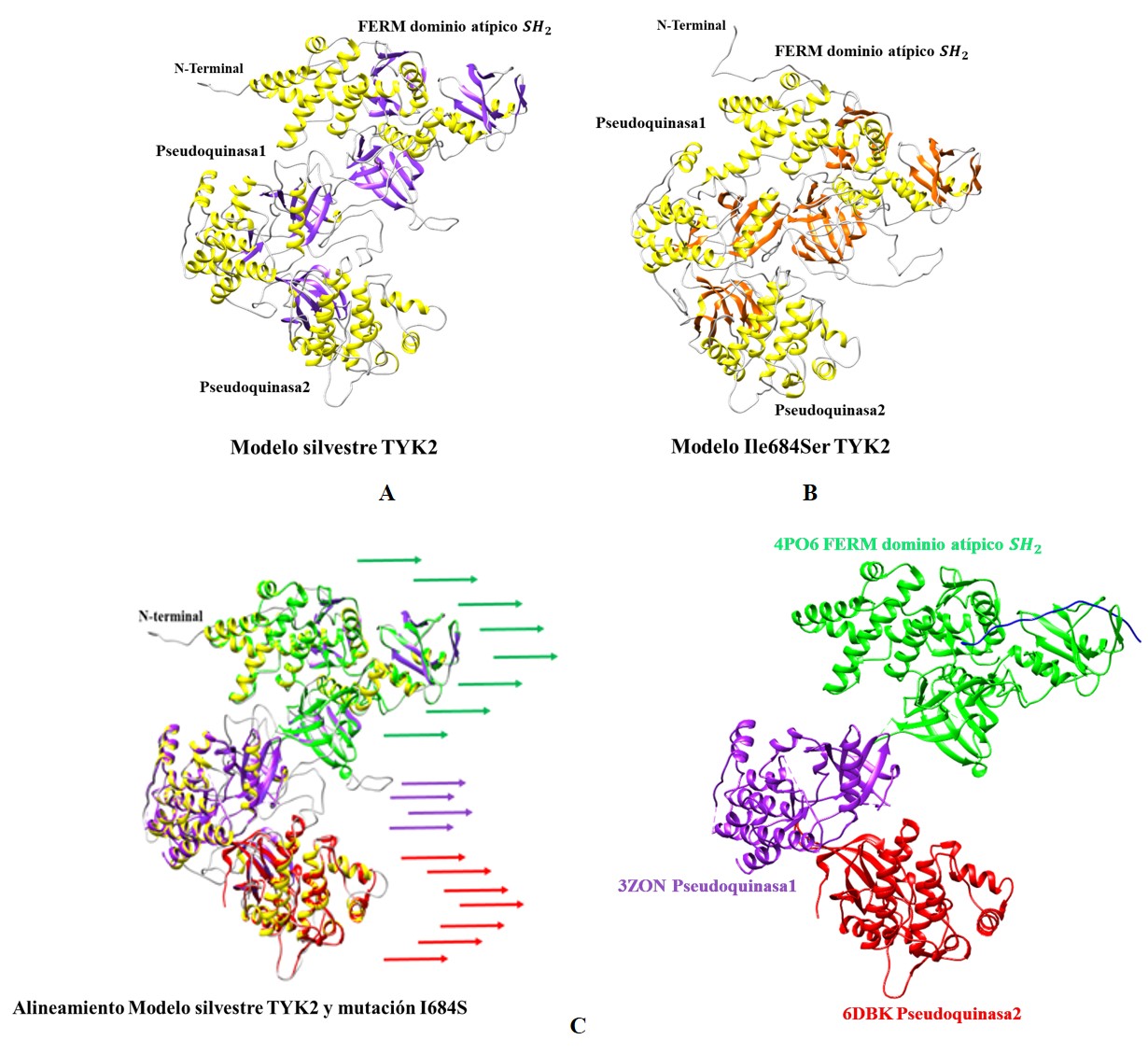
La psoriasis es una enfermedad inflamatoria crónica inmunomediada en la que el eje IL-23/Th17 dirige el proceso patogénico. En la regulación del sistema inmunitario la propensión al desarrollo de la psoriasis en una población colombiana se asoció con la variante no sinónima I684S en TYK2. En el estudio funcional del efecto de la variante a nivel molecular, se hizo la modelación molecular de la enzima a partir de su secuencia primaria, lo que permite reconocer el plegamiento tridimensional y unificar así el modelo con las estructuras cristalizadas de las diferentes subunidades de la enzima que han sido reportadas como aisladas y sin conexión. Este modelo final se refinó utilizando dos programas y se seleccionó el mejor para la simulación posterior según la puntuación de los valores energéticos y estereoquímicos. La simulación molecular se hizo con el método semiempírico del programa Spartan 18’ y el campo de fuerza del Austin Model 1 para la región vecinal que incluye la posición 684 en donde se hizo el cálculo energético y de superficie electrónica bajo un enfoque mecánico cuántico. Los resultados evidenciaron que los modelos presentaron cambios tridimensionales a nivel topológico de la enzima TYK2 silvestre y la mutación I684S en los patrones del enlace de hidrógeno y en la estructura electrónica, con un aumento de la densidad electrónica en la mutación que favorece la fosforilación de quinasas en la transducción de la señal y la desregulación inmunológica.
Referencias
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., Duvaud, S., Flegel, V., Fortier, A., Gasteiger, E., Grosdidier, A., Hernandez, C., Ioannidis, V., Kuznetsov, D., Liechti, R., Moretti, S., Mostaguir, K., Redaschi, N., Rossier, G., …
Stockinger, H. (2012). ExPASy: SIB bioinformatics resource portal. Nucleic Acids Research, 40(W1), 597-603. https://doi.org/10.1093/nar/gks400
Benkert, P., Tosatto, S. C. E., Schomburg, D. (2008). QMEAN: A comprehensive scoring function for model quality assessment. Proteins: Structure, Function, and Bioinformatics, 71(1), 261-277. https://doi.org/10.1002/prot.21715
Berman, H. M., Kleywegt, G. J., Nakamura, H., Markley, J. L. (2012). The protein data bank at 40: Reflecting on the past to prepare for the future. Structure, 20(3), 391–396. https://doi.org/10.1016/j.str.2012.01.010
Blom, N., Sicheritz-Pontén, T., Gupta, R., Gammeltoft, S., Brunak, S. (2004). Prediction of posttranslational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics, 4(6), 1633-1649. https://doi.org/10.1002/pmic.200300771
Boehncke, W.-H. & Schön, M. P. (2015). Psoriasis. The Lancet, 386(9997), 983–994. https://doi.org/10.1016/S0140-6736(14)61909-7
Cano, L., Soto-Ospina, A., Araque, P., Caro-Gómez, M.A. (2021). Diffusion Mechanism Modeling of Metformin in Human Organic Cationic Amino Acid Transporter one and Functional Impact of S189L , R206C , and G401S Mutation. Frontiers in Pharmacology, 11, 1–14. https://doi.org/10.3389/fphar.2020.587590
Cardona-Pemberthy, V., Rendón, M., Beltrán, J. C., Soto-Ospina, A., Muñoz-Gomez, A., Araque-Marín, P., Corredor, M., Bedoya, G., Cardona-Castro, N. (2018). Genetic variants, structural, and functional changes of Myelin Protein Zero and Mannose-Binding Lectin 2 protein involved in immune response and its allelic transmission in families of patients with leprosy in Colombia. Infection, Genetics and Evolution, 61(March), 215-223.https://doi.org/10.1016/j.meegid.2018.04.002
Carter, M., Jemth, A. S., Hagenkort, A., Page, B. D. G., Gustafsson, R., Griese, J. J., Gad, H., Valerie, N. C. K., Desroses, M., Boström, J., Warpman Berglund, U., Helleday, T., Stenmark, P. (2015). Crystal structure, biochemical and cellular activities demonstrate separate functions of MTH1 and MTH2. Nature Communications, 6, 4–13. https://doi.org/10.1038/ncomms8871
Chandran, V. (2013). The Genetics of Psoriasis and Psoriatic Arthritis. Clinical Reviews in Allergy & Immunology, 44(2), 149-156. https://doi.org/10.1007/s12016-012-8303-5
Crystallography and Bioinformatics Group. (2017). Rampage: Ramachandran plot. http://mordred.bioc.cam.ac.uk/~rapper/rampage.php
Dewar, M.J., Zoebisch, E.G., Healy, E.F., Stewart, J. P. (1993). AM1: A Quantum Mechanical Molecular Model. J. Am. Chem. Soc., 49(June), 3903-3909.
Elkins, J.M., Wang, J., Krojer, T., Savitsky, P., Chalk, R., Daga, N., Salah, E., Berridge, G., Picaud, S., von Delft, F., Bountra, C., Edwards, A., Knapp, S. (2013). Human TYK2 pseudokinase domain bound to a kinase inhibitor. Protein Data Bank Europe. https://doi.org/10.2210/pdb3ZON/pdb
Ellinghaus, D., Jostins, L., Spain, S. L., Cortes, A., Bethune, J., Han, B., Park, Y. R., Raychaudhuri, S., Pouget, J. G., Hübenthal, M., Folseraas, T., Wang, Y., Esko, T., Metspalu, A., Westra, H.-J., Franke, L., Pers, T. H., Weersma, R. K., Collij, V., … Franke, A. (2016). Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights diseasespecific patterns at shared loci. Nature Genetics, 48(5), 510. https://doi.org/10.1038/NG.3528
Enerbäck, C., Sandin, C., Lambert, S., Zawistowski, M., Stuart, P. E., Verma, D., Tsoi, L.C., Nair, R. P., Johnston, A., Elder, J.T. (2018). The psoriasis-protective TYK2 I684S variant impairs IL-12 stimulated pSTAT4 response in skin-homing CD4+ and CD8+ memory T-cells. Scientific Reports, 8(1), 7043. https://doi.org/10.1038/s41598-018-25282-2
Farber, E.M. & Nall, L. (1994). Psoriasis in the tropics. Epidemiologic, genetic, clinical, and therapeutic aspects. Dermatologic Clinics, 12(4), 805-816.
Fensome, A., Ambler, C. M., Arnold, E., Banker, M. E., Brown, M. F., Chrencik, J., Clark, J. D., Dowty, M. E., Efremov, I. V., Flick, A., Gerstenberger, B. S., Gopalsamy, A., Hayward, M. M., Hegen, M., Hollingshead, B. D., Jussif, J., Knafels, J. D., Limburg, D. C., Lin, D., … Zhang, L. (2018). Dual Inhibition of TYK2 and JAK1 for the Treatment of Autoimmune Diseases: Discovery of ((S)-2,2-Difluorocyclopropyl)((1 R,5 S)-3-(2-((1-methyl-1 H-pyrazol-4-yl)amino)pyrimidin-4-yl)-3,8-diazabicyclo[3.2.1]octan-8-yl) methanone (PF-06700841). Journal of Medicinal Chemistry, 61(19), 8597-8612. https://doi.org/10.1021/acs.jmedchem.8b00917
Foresman, J. & Frisch, E. (1996). Exploring chemistry with electronic structure methods, Gaussian Inc. (ed.); Second edition. Gaussian Inc.
Freeman, A. K., Linowski, G. J., Brady, C., Lind, L., VanVeldhuisen, P., Singer, G., Lebwohl, M. (2003). Tacrolimus ointment for the treatment of psoriasis on the face and intertriginous areas. Journal of the American Academy of Dermatology, 48(4), 564-568. https://doi.org/10.1067/mjd.2003.169
Ghoreschi, K., Augustin, M., Baraliakos, X., Krönke, G., Schneider, M., Schreiber, S., Schulze-Koops, H., Zeißig, S., Thaçi, D. (2021). TYK2 inhibition and its potential in the treatment of chronic inflammatory immune diseases. JDDG - Journal of the German Society of Dermatology. https://doi.org/10.1111/ddg.14585
Griffiths, C.E.M. (2003). The immunological basis of psoriasis. Journal of the European Academy of Dermatology and Venereology:JEADV, 17 Suppl 2, 1-5.
Grillo, I.B., Urquiza-Carvalho, G. A., Bachega, J.F.R., Rocha, G.B. (2020). Elucidating Enzymatic Catalysis Using Fast Quantum Chemical Descriptors. Journal of Chemical Information and Modeling, 60(2), 578-591. https://doi.org/10.1021/acs.jcim.9b00860
Grjibovski, A., Olsen, A., Magnus, P., Harris, J. (2007). Psoriasis in Norwegian twins: contribution of genetic and environmental effects. Journal of the European Academy of Dermatology and Venereology, 21(10), 1337-1343. https://doi.org/10.1111/j.1468-3083.2007.02268.x
Gudjonsson, J.E. & Elder, J.T. (2007). Psoriasis: epidemiology. Clinics in Dermatology, 25(6), 535-546. https://doi.org/10.1016/J.CLINDERMATOL.2007.08.007
Guex, N. & Peitsch, M.C. (1997). SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis, 18(15), 2714-2723. https://doi.org/10.1002/elps.1150181505
Kelley, L.A., Mezulis, S., Yates, C., Wass, M., Sternberg, M. (2015). The Phyre2 web portal for protein modelling, prediction, and analysis. Nature Protocols, 10(6), 845-858. https://doi.org/10.1038/nprot.2015-053
Kelley, L.A. & Sternberg, M.J.E. (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nature Protocols, 4(3), 363-371. https://doi.org/http://dx.doi.org/10.1038/nprot.2009.2
Kisiel, B., Kisiel, K., Szymański, K., Mackiewicz, W., Biało-Wó Jcicka, E., Uczniak, S., Fogtman, A., Iwanicka-Nowicka, R., Koblowska, M., Kossowska, H., Placha, G., Sykulski, M., Bachta, A., Tłustochowicz, W., Płoski, R., Kaszuba, A. (2017). The association between 38 previously reported polymorphisms and psoriasis in a Polish population: High predicative accuracy of a genetic risk score combining 16 loci. PLoS ONE 12(6): e0179348. https://doi.org/10.1371/journal.pone.0179348
Kurd, S.K. & Gelfand, J.M. (2009). The prevalence of previously diagnosed and undiagnosed psoriasis in US adults: Results from NHANES 2003-2004. Journal of the American Academy of Dermatology, 60(2), 218-224. https://doi.org/10.1016/j.jaad.2008.09.022
Langley, R.G. & Ellis, C.N. (2004). Evaluating psoriasis with psoriasis area and severity index, psoriasis global assessment, and lattice system physician’s global assessment. Journal of the American Academy of Dermatology, 51(4), 563-569.
Lee, Y.-A., Rüschendorf, F., Windemuth, C., Schmitt-Egenolf, M., Stadelmann, A., Nürnberg, G., Ständer, M., Wienker, T.F., Reis, A., Traupe, H. (2000). Genomewide Scan in German Families Reveals Evidence for a Novel Psoriasis-Susceptibility Locus on Chromosome 19p13. The American Journal of Human Genetics, 67(4), 1020-1024. https://doi.org/10.1086/303075
Li, Z., Gakovic, M., Ragimbeau, J., Eloranta, M.-L., Ronnblom, L., Michel, F., Pellegrini, S. (2013). Two Rare Disease-Associated Tyk2 Variants Are Catalytically Impaired but Signaling Competent. The Journal of Immunology, 190(5), 2335-2344. https://doi.org/10.4049/
jimmunol.1203118
Li, Zhi, Gakovic, M., Ragimbeau, J., Eloranta, M.-L., Rönnblom, L., Michel, F., Pellegrini, S. (2013). Two Rare Disease-Associated Tyk2 Variants Are Catalytically Impaired but Signaling Competent. The Journal of Immunology, 190(5), 2335-2344. https://doi.org/10.4049/
jimmunol.1203118
Lønnberg, A. S., Skov, L., Skytthe, A., Kyvik, K.O., Pedersen, O.B., Thomsen, S.F. (2013). Heritability of psoriasis in a large twin sample. British Journal of Dermatology, 169(2), 412-416. https://doi.org/10.1111/bjd.12375
Lønnberg, A., Skov, L., Duffy, D., Skytthe, A., Kyvik, K., Pedersen, O., Thomsen, S. (2016). Genetic Factors Explain Variation in the Age at Onset of Psoriasis: A Population-based Twin Study. Acta Dermato Venereologica, 96(1), 35–38. https://doi.org/10.2340/00015555-2171
López-Rivera, J.J., Rodríguez-Salazar, L., Soto-Ospina, A., Estrada-Serrato, C., Serrano, D., Chaparro-Solano, H.M., Londoño, O., Rueda, P.A., Ardila, G., Villegas-Lanau, A., Godoy-Corredor, M., Cuartas, M., Vélez, J. I., Vidal, O.M., Isaza-Ruget, M.A., Arcos-Burgos, M. (2022). Structural Protein Effects Underpinning Cognitive Developmental Delay of the PURA p.Phe233del Mutation Modelled by Artificial Intelligence and the Hybrid Quantum Mechanics–Molecular Mechanics Framework. Brain Sciences, 12(7). https://doi.org/10.3390/brainsci12070871
Luger, T.A. & Loser, K. (2018). Novel insights into the pathogenesis of psoriasis. Clinical Immunology, 186, 43-45. https://doi.org/10.1016/j.clim.2017.07.014
Mahil, S.K., Capon, F., Barker, J.N. (2015). Genetics of Psoriasis. Dermatologic Clinics, 33(1),1-11. https://doi.org/10.1016J.DET.2014.09.001
Maniatis, S.E.F.T. (1989). Genetics of Psoriasis and Pharmacogenetics of Biological Drugs. In Autoimmune Diseases (2nd ed., Vol. 2013). https://doi.org/10.1155/2013/613086
Min, X., Ungureanu, D., Maxwell, S., Hammarén, H., Thibault, S., Hillert, E. K., Ayres, M., Greenfield, B., Eksterowicz, J., Gabel, C., Walker, N., Silvennoinen, O., Wang, Z. (2015). Structural and functional characterization of the JH2 pseudokinase domain of JAK family tyrosine kinase 2 (TYK2). Journal of Biological Chemistry, 290(45), 27261-27270. https://doi.org/10.1074/jbc.M115.672048
Nititham, J., Gupta, R., Zeng, X., Hartogensis, W., Nixon, D. F., Deeks, S.G., Hecht, F.M., Liao, W. (2017). Psoriasis risk SNPs and their association with HIV-1 control. Human Immunology, 78(2), 179. https://doi.org/10.1016/J.HUMIMM.2016.10.018
Peng, C., Wang, J., Yu, Y., Wang, G., Chen, Z., Xu, Z., Cai, T., Shao, Q., Shi, J., Zhu, W. (2019). Improving the accuracy of predicting protein-ligand binding-free energy with semiempirical quantum chemistry charge. Future Medicinal Chemistry, 11(4), 303-321. https://doi.org/10.4155/fmc-2018-0207
Pettersen, E.F., Goddard, T.D., Huang, C.C., Couch, G.S., Greenblatt, D.M., Meng, E.C., Ferrin, T.E. (2004). UCSF Chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605-1612. https://doi.org/10.1002/
jcc.20084
Prieto-Pérez, R., Solano-López, G., Cabaleiro, T., Román, M., Ochoa, D., Talegón, M., Baniandrés, O., López-Estebaranz, J. L., Cueva, P. de la, Daudén, E., Abad-Santos, F. (2015). Polymorphisms Associated with Age at Onset in Patients with Moderate-to-Severe Plaque Psoriasis. Journal of Immunology Research, 2015, Article ID 101879, 8 pages. https://doi.org/10.1155/2015/101879
Rahman, P. & Elder, J.T. (2005). Genetic epidemiology of psoriasis and psoriatic arthritis. Annals of the rheumatic diseases, 64(suppl 2), ii37-ii39.
Soto-Ospina, A., Araque Marín, P., Bedoya, G. D. J., Villegas Lanau, A. (2021a). Structural Predictive Model of Presenilin-2 Protein and Analysis of Structural Effects of Familial Alzheimer’s Disease Mutations. Biochemistry Research International, 2021, Article ID 9542038, 20 pages. https://doi.org/10.1155/2021/9542038
Soto-Ospina, A., Araque Marín, P., Bedoya, G., Sepulveda-Falla, D., Villegas Lanau, A. (2021b). Protein Predictive Modeling and Simulation of Mutations of Presenilin-1 Familial Alzheimer’s Disease on the Orthosteric Site. Frontiers in Molecular Biosciences, 8(June), 1-15. https://doi.org/10.3389/fmolb.2021.649990
Strange, A., Capon, F., Spencer, C.C.A., Knight, J., Weale, M. E., Allen, M. H., Barton, A., Band, G., Bellenguez, C., Bergboer, J. G. M., Blackwell, J. M., Bramon, E., Bumpstead, S. J., Casas, J. P., Cork, M. J., Corvin, A., Deloukas, P., Dilthey, A., Duncanson, A., … Trembath, R.C. (2010). A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nature Genetics, 42(11), 985-990. https://doi.org/10.1038/ng.694
Tejada-Moreno, J.A., Villegas-Lanau, A., Madrigal-Zapata, L., Baena-Pineda, A. Y., Vélez- Hernández, J., Campo-Nieto, O., Soto-Ospina, A., Araque-Marín, P., Rishishwar, L., Norris, E. T., Chande, A. T., Jordan, I. K., Berrío, G.B. (2022). Mutations in SORL1 and MTHFDL1 possibly contribute to the development of Alzheimer’s disease in a multigenerational Colombian Family. PLoS ONE, 17(7 July), 1-28. https://doi.org/10.1371/journal.pone.0269955
Wallweber, H.J.A., Tam, C., Franke, Y., Starovasnik, M.A., Lupardus, P. J. (2014). Structural basis of recognition of interferon-α receptor by tyrosine kinase 2. Nature Structural and Molecular Biology, 21(5), 443–448. https://doi.org/10.1038/nsmb.2807
Wavefunction. (1991). Spartan 20’. Wavefunction. https://www.wavefun.com/corporate/more_spartan.html
Xu, D. & Zhang, Y. (2011). Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophysical Journal, 101(10), 2525-2534. https://doi.org/10.1016/j.bpj.2011.10.024
Zhang, J., Liang, Y., Zhang, Y. (2011). Atomic-level protein structure refinement using fragmentguided molecular dynamics conformation sampling. Structure, 19(12), 1784-1795. https://doi.org/10.1016/j.str.2011.09.022
Zhao, Y., Soh, T. S., Zheng, J., Chan, K. W. K., Phoo, W. W., Lee, C. C., Tay, M. Y. F., Swaminathan, K., Cornvik, T. C., Lim, S. P., Shi, P. Y., Lescar, J., Vasudevan, S. G., Luo, D. (2015). A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Interdomain Interface Essential for Protein Flexibility and Virus Replication. PLoS Pathogens, 11(3), 1-27. https://doi.org/10.1371/journal.ppat.1004682

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-SinDerivadas 4.0.
Derechos de autor 2023 Revista de la Academia Colombiana de Ciencias Exactas, Físicas y Naturales