Resumen
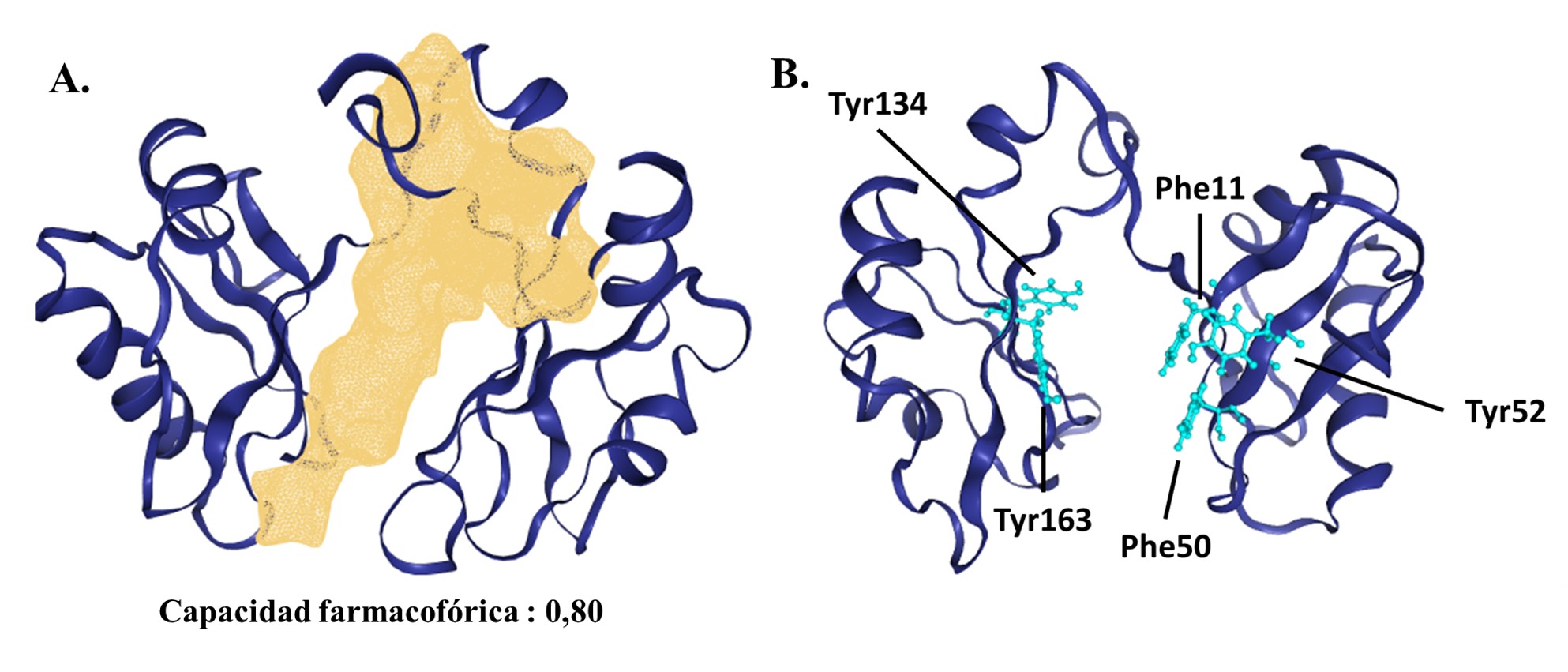
Varios estudios proponen la nucleolina como un blanco alternativo para la búsqueda y el diseño de nuevas terapias antitumorales o anticancerígenas. El objetivo de este estudio fue evaluar compuestos con potenciales afinidades de unión al ARN de las nucleolinas mediante herramientas bioinformáticas. Se evaluaron diez compuestos de los cuales tres moléculas (ácido betulínico, triptolide y berberina) presentaron afinidades de interacción con el dominio de unión al ARN (RNA binding domain, RBD), exhibiendo valores de energía de unión significativamente favorables (p<0,05) y una constante de disociación calculada (Kd calc) entre 0,14 y 0,91 μM. Las simulaciones de dinámica molecular evidenciaron que únicamente el complejo nucleolina-berberina tuvo estabilidad de interacción y energía libre de unión favorable en el tiempo simulado. Los residuos involucrados en la formación del complejo eran aminoácidos que cumplen funciones importantes en el sitio activo. Los hallazgos sugieren que, entre los compuestos evaluados, la berberina tuvo resultados favorables como potencial inhibidor de las actividades de la nucleolina, específicamente del dominio RBD.
Referencias
Allain F. H. T., Gilbert D. E., Bouvet P., Feigon J. (2000). Solution structure of the two N-terminal RNA-binding domains of nucleolin and NMR study of the interaction with its RNA target. Journal of Molecular Biology, 303(2), 227-241. https://doi.org/10.1006/jmbi.2000.4118
Arumugam S., Clarke Miller M., Maliekal J., Bates P. J., Trent J. O., Lane A. N. (2010). Solution structure of the RBD 1, 2 domains from human nucleolin. Journal of Biomolecular NMR, 47(1), 79-83. https://doi.org/10.1007/s10858-010-9412-1
Åqvist, J., Medina, C., Samuelsson, J.E. (1994). A new method for predicting binding affinity in computer-aided drug design. Protein Engineering, Design and Selection, 7(3), 385-391. https://doi.org/10.1093/protein/7.3.385
Berendsen, H. J., Postma, J. V., Van Gunsteren, W. F., DiNola, A. R. H. J., Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684-3690. https://doi.org/10.1063/1.448118
Berger C. M., Gaume X., Bouvet P. (2015). The roles of nucleolin subcellular localization in cancer. Biochimie, 113, 78-85. https://doi.org/10.1016/j.biochi.2015.03.023
Berman H. M., Westbrook J., Feng Z., Gilliland G., Bhat T. N., Weissig H., Shindyalov I. N., Bourne P. E. (2000). The Protein Data Bank. Nucleic Acids Research, 28(1), 235-242. https://doi.org/10.1093/nar/28.1.235
Berrada M., Serreqi A., Dabbarh F., Owusu A., Gupta A., Lehnert S. (2005). A novel non-toxic camptothecin formulation for cancer chemotherapy. Biomaterials, 26(14), 2115-2120. https://doi.org/10.1016/j.biomaterials.2004.06.013
Biao L., Qihou W., Jin L., Shiqing H., Bing H. (2007). Study of the effect of berberine on the telomerase activity of human nasopharyngeal carcinoma (NPC) CNE-2. Shanxi Medical Journal, 36, 1281-1283.
Bissantz, C., Kuhn, B., Stahl, M. (2010). A medicinal chemist’s guide to molecular interactions. Journal of Medicinal Chemistry, 53(14), 5061-5084. https://doi.org/10.1021/jm100112j
Bouvet P., Allain F. H. T., Finger L. D., Dieckmann T., Feigon, J. (2001). Recognition of pre-formed and flexible elements of an RNA stem-loop by nucleolin. Journal of Molecular Biology, 309 (3), 763-775. https://doi.org/10.1006/jmbi.2001.4691
Böhm, H. J., Klebe, G. (1996). What can we learn from molecular recognition in protein–ligand complexes for the design of new drugs? Angewandte Chemie 35(22), 2588-2614. https://doi.org/10.1002/anie.199625881
Brooks B. R., Brooks C. L., Mackerell A. D., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., Fischer S., Gao J., Hodoscek M., Im W., Lazaridis T., Ma J., Ovchinnikov V., Paci E., Pastor R. W., Post C. B., Pu J. Z., Schaefer M., Tidor, B., Venable R. W., Woodcock H. L., Wu X., Yang W., York D. M., Karplus M. (2009). CHARMM: The biomolecular simulation program. Journal of Computational Chemistry, 30(10), 1545-1614. https://doi.org/10.1002/jcc.21287
Bugler B., Caizergues-Ferrer M., Bouche G., Bourbon H., Amalric F. (1982). Detection and localization of a class of proteins immunologically related to a 100‐KDa nucleolar protein. European Journal of Biochemistry, 128(2‐3), 475-480. https://doi.org/10.1111/j.1432-1033.1982.tb06989.x
Chen Z., Xu X. (2016). Roles of nucleolin: Focus on cancer and anti-cancer therapy. Saudi Medical Journal, 37(12), 1312-1318. https://doi.org/10.15537/smj.2016.12.15972
Chintharlapalli S., Papineni S., Ramaiah S. K., Safe S. (2007). Betulinic acid inhibits prostate cancer growth through inhibition of specificity protein transcription factors. Cancer Research, 67(6), 2816-2823. https://doi.org/10.1158/0008-5472.CAN-06-3735
Choudhury A., Das N. C., Patra R., Bhattacharya M., Ghosh P., Patra B. C., Mukherjee S. (2021). Exploring the binding efficacy of ivermectin against the key proteins of SARS-CoV-2 pathogenesis: an in silico approach. Future Virology, 16 (4), 277-291. https://doi.org/10.2217/fvl-2020-0342
Daina A., Zoete V. (2016). A BOI-LED-Egg to Predict Gastrointestinal Absorption and Brain Penetration of Small Molecules. Chemi Med Chem, 11(11), 1117-1121. https://doi.org/10.1002/cmdc.20160018
Daina A., Michielin O., Zoete V. (2014). iLOGP: A Simple, Robust, and Efficient Description of n-Octanol/Water Partition Coefficient for Drug Design Using the GB/SA Approach. Journal of Chemical Information and Modeling, 54(12), 3284-3301. https://doi.org/10.1021/
ci500467k
Daina A., Michielin O., Zoete V. (2017). SwissADME: A free web tool to evaluate pharmaco-kinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Scientific Reports, 7(1), 42717. https://doi.org/10.1038/srep42717
Dallakyan S., Olson A. J. (2015). Small Molecule Library Screening by Docking with PyRx. En J. E. Hempel, C. H. Williams, & C. C. Hong (Eds.), Chemical Biology: Methods and Protocols (pp. 243-250). Springer. https://doi.org/10.1007/978-1-4939-2269-7_19
Dassault Systèmes BIOVIA. (2019). Discovery Studio Visualizer, version 20.1, San Diego: Dassault Systèmes.
De Amorim H. L. N., Cáceres R. A., Netz P. A. (2008). Linear interaction energy (LIE) method in lead discovery and optimization. Current Drug Targets, 9(12), 1100-1105. https://doi.org/10.2174/138945008786949360
Dunn O. J. (1964). Multiple Comparisons Using Rank Sums. Technometrics, 6(3), 241-252. https://doi.org/10.1080/00401706.1964.10490181
Ginisty H., Sicard H., Roger B., Bouvet P. (1999). Structure and functions of nucleolin. Journal of Cell Science, 112(6), 761-772. https://doi.org/10.1242/jcs.112.6.761
Ginisty H., Amalric F., Bouvet P. (2001). Two different combinations of RNA-binding domains determine the RNA binding specificity of nucleolin. Journal of Biological Chemistry, 276(17), 14338-14343. https://doi.org/10.1074/jbc.M011120200
Hammer O., Harper D. A. T., Ryan P. D. (2001). PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica, 4(1), 9.
Hansson T., Marelius J., Åqvist J. (1998). Ligand binding affinity prediction by linear interaction energy methods. Journal of Computer-Aided Molecular Design, 12(1), 27-35. https://doi.org/10.1023/A:1007930623000
Hanwell M. D., Curtis D. E., Lonie D. C., Vandermeersch T., Zurek E., Hutchison G. R. (2012). Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. Journal of Cheminformatics, 4(1), 17. https://doi.org/10.1186/1758-2946-4-17
Hopkins A. L., Keserü, G. M., Leeson P. D., Rees D. C., Reynolds C. H. (2014). The role of ligand efficiency metrics in drug discovery. Nature Reviews Drug Discovery, 13(2), 105-121. https://doi.org/10.1038/nrd4163
Hou W., Liu B., Xu H. (2019). Triptolide: Medicinal chemistry, chemical biology and clinical progress. European Journal of Medicinal Chemistry, 176, 378-392. https://doi.org/ 10.1016/j.ejmech.2019.05.032
Howat S., Park B., Oh I. S., Jin Y. W., Lee E. K., Loake G. J. (2014). Paclitaxel: biosynthesis, production and future prospects. New Biotechnology, 31(3), 242-245. https://doi.org/10.1016/j.nbt.2014.02.010
Hsu W. H., Hsieh Y. S., Kuo H. C., Teng C. Y., Huang H. I., Wang C. J., Yang S. F., Liou Y. S., Kuo W. H. (2007). Berberine induces apoptosis in SW620 human colonic carcinoma cells through generation of reactive oxygen species and activation of JNK/p38 MAPK and FasL. Archives of Toxicology, 81(10), 719-728. https://doi.org/10.1007/s00204-006-0169-y
Humphrey W., Dalke A., Schulten K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33-38. https://doi.org/10.1016/0263-7855(96)00018-5
Imanshahidi M. & Hosseinzadeh H. (2008). Pharmacological and therapeutic effects of Berberis vulgaris and its active constituent, berberine. Phytotherapy Research, 22(8), 999-1012. https://doi.org/10.1002/ptr.2399
Islam, M. M., Kumar G. S. (2008). RNA targeting by small molecule alkaloids: studies on the binding of berberine and palmatine to polyribonucleotides and comparison to ethidium. Journal of Molecular Structure, 875(1-3), 382-391. https://doi.org/10.1016/j.molstruc.2007.05.004
Jabbarzadeh Kaboli P., Leong M. P. Y., Ismail P., Ling K. H. (2019). Antitumor effects of berberine against EGFR, ERK1/2, P38 and AKT in MDA-MB231 and MCF-7 breast cancer cells using molecular modelling and in vitro study. Pharmacological Reports, 71(1), 13-23. https://doi.org/10.1016/j.pharep.2018.07.005
Jo S., Kim T., Iyer V. G., Im W. (2008). CHARMM‐GUI: a web‐based graphical user interface for CHARMM. Journal of Computational Chemistry, 29(11), 1859-1865. https://doi.org/10.1002/jcc.20945
Johnson L., Swie Goping I., Rieger A., Mane J., Huzil T., Banerjee A., Luduena R., Hassani B., Winter P., Tuszynski A. J. (2017). Novel colchicine derivatives and their anti-cancer activity. Current Topics in Medicinal Chemistry, 17(22), 2538-2558. https://doi.org/10.2174/
Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926-935. https://doi.org/10.1063/1.445869
Khazir J., Mir B. A., Pilcher L., Riley D. L. (2014). Role of plants in anticancer drug discovery. Phytochemistry Letters, 7, 173-181. https://doi.org/10.1016/j.phytol.2013.11.010
Kim J. S., Oh D., Yim M. J., Park J. J., Kang K. R., Cho I. A., Moon S. M., Oh J. S., You J. S., Kim C. S., Kim D. K., Lee S. Y., Lee G. J., Im H. J., Kim S. G. (2015). Berberine induces FasL-related apoptosis through p38 activation in KB human oral cancer cells. Oncology Reports, 33(4), 1775-1782. https://doi.org/10.3892/or.2015.3768
Kim S., Lee J., Jo S., Brooks III C. L., Lee H. S., Im W. (2017). CHARMM‐GUI ligand reader and modeler for CHARMM force field generation of small molecules. Journal of Computational Chemistry, 38, 1879-1886. https://doi.org/10.1002/jcc.24829
Kim S., Thiessen P. A., Bolton E. E., Chen J., Fu G., Gindulyte A., Han L., He J., He S., Shoemaker, B. A., Wang J., Yu B., Zhang J., Bryant S. H. (2016). PubChem Substance and Compound data-bases. Nucleic Acids Research, 44(D1), D1202-D1213. https://doi.org/10.1093/nar/gkv951
Kruskal W. H. & Wallis W. A. (1952). Use of Ranks in One-Criterion Variance Analysis. Journal of the American Statistical Association, 47(260), 583-621. https://doi.org/10.1080/01621459.1952.10483441
Lee J., Cheng X., Swails J. M., Yeom M. S., Eastman P. K., Lemkul J. A., Wei S., Buckner J., Jeong J. C., Qi Y., Jo S., Pande V. S., Case D. A., Brooks C. L., Mac-Kerell A. D., KlaudaJ. B., Im W. (2016). CHARMM-GUI Input Generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM Simulations Using the CHARMM36 Additive Force Field. Journal of Chemical Theory and Computation, 12(1), 405-413. https://doi.org/10.1021/acs.jctc.5b00935
Lin S. S., Chung J. G., Lin J. P., Chuang J. Y., Chang W. C., Wu J. Y., Tyan Y. S. (2005). Berberine inhibits arylamine N-acetyltransferase activity and gene expression in mouse leukemia L 1210 cells. Phytomedicine, 12(5), 351-358. https://doi.org/10.1016/j.phymed.2003.11.008
Lipinski, C. A., Lombardo, F., Dominy, B. W., Feeney, P. J. (2001). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development. Advanced Drug Delivery Reviews, 46, 3-26. https://doi.org/10.1016/S0169-409X(00)00129-0
Liu, X., Wang, K., Duan, N., Lan, Y., Ma, P., Zheng, H., Li, J., Hua, Z. C. (2015). Computational prediction and experimental validation of low-affinity target of triptolide and its analogues. RSC advances, 5(44), 34572-34579. https://doi.org/10.1039/C4RA17009A
Liu, H., Hou, T. (2016). CaFE: A tool for binding affinity prediction using end-point free energy methods. Bioinformatics, 32(14), 2216-2218. https://doi.org/10.1093/bioinformatics/btw215 Liu, H., Shen, M., Zhao, D., Ru, D., Duan, Y., Ding, C., Li, H. (2019). The effect of triptolide-loaded exosomes on the proliferation and apoptosis of human ovarian cancer SKOV3 cells. BioMed Research International, 2019, 1-14. https://doi.org/10.1155/2019/2595801
Losfeld M. E., El Khoury D., Mariot P., Carpentier M., Krust B., Briand J. P., Mazurier J., Hovanessian A. G., Legrand, D. (2009). The cell surface expressed nucleolin is a glycoprotein that triggers calcium entry into mammalian cells. Experimental Cell Research, 315(2), 357-369. https://doi.org/10.1016/j.yexcr.2008.10.039
Meng G. Z., Xiao S. J., Zeng S. E., Li Y. Q. (2011). Downregulation of cell-surface-expressed nucleolin inhibits the growth of hepatocellular carcinoma cells in vitro. Zhonghua zhong liu za zhi [Chinese journal of oncology], 33 (1), 23-27.
Onawole A. T., Kolapo T. U., Sulaiman K. O., Adegoke R. O. (2018). Structure based virtual screening of the Ebola virus trimeric glycoprotein using consensus scoring. Computational Biology and Chemistry, 72, 170-180. https://doi.org/10.1016 j.compbiolchem.2017.11.006
Palmieri A., Scapoli L., Iapichino A., Mercolini L., Mandrone M., Poli F., Giannì A. B., Baserga C., Martinelli M. (2019). Berberine and Tinospora cordifolia exert a potential anticancer effect on colon cancer cells by acting on specific pathways. International Journal of Immunopathology and Pharmacology, 33, 1-10. https://doi.org/10.1177/2058738419855567
Pan L., Chai H. B., Kinghorn A. D. (2012). Discovery of new anticancer agents from higher plants. Frontiers in Bioscience (Schol Ed), 4(1), 142-156. https://doi.org/10.2741/s257
Parchehbaf-Jadid A., Zarefatin L., Javadi L. (2012). A theoretical study on interactions between Berberine as an anticancer drug and DNA. Journal of the Iranian Chemical Research, 5(3), 143-153.
Parrinello, M., Rahman, A. (1980). Crystal structure and pair potentials: A molecular-dynamics study. Physical Review Letters, 45(14), 1196-1199. https://doi.org/10.1103/PhysRevLett.45.1196
Patil, R., Das, S., Stanley, A., Yadav, L., Sudhakar, A., Varma, A. K. (2010). Optimized hydro-phobic interactions and hydrogen bonding at the target-ligand interface leads the pathways of drug-designing. PloS one, 5(8), e12029. https://doi.org/10.1371/journal.pone.0012029
Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005). Scalable molecular dynamics with NAMD. Journal of Computational Chemistry, 26(16), 1781-1802. https://doi.org/10.1002/jcc.20289
Pisha E., Chai H., Lee I. S., Chagwedera T. E., Farnsworth N. R., Cordell G. A., Beecher C. W. W., Fong H. H. S., Kinghorn A. D., Brown D. M., Wani M. C., Wall M. E., Hieken T. J., Das Gupta T. K., Pezzuto J. M. (1995). Discovery of betulinic acid as a selective inhibitor of human melanoma that functions by induction of apoptosis. Nature Medicine, 1(10), 1046-1051. https://doi.org/10.1038/nm1095-1046
Qin Y., Pang J. Y., Chen W. H., Zhao Z. Z., Liu L., Jiang Z. H. (2007). Inhibition of DNA topoisomerase I by natural and synthetic mono‐and dimeric protoberberine alkaloids. Chemistry & Biodiversity, 4(3), 481-487. https://doi.org/10.1002/cbdv.200790040
Rauf A., Abu-Izneid T., Khalil A. A., Imran M., Shah Z. A., Emran T. B., Mitra S., Khan Z., Alhumaydhi F. A., Aljohani A. S. M., Khan I., Rahman M., Jeandet P., Gondal T. A. (2021). Berberine as a potential anticancer agent: A comprehensive review. Molecules, 26(23), 7368. https://doi.org/10.3390/molecules26237368
Santana C. A., Silveira S. D. A., Moraes J. P., Izidoro S. C., de Melo-Minardi R. C., Ribeiro A. J., Tyzack J. D. N., Thornton J. M. (2020). GRaSP: a graph-based residue neighborhood strategy to predict binding sites. Bioinformatics, 36(26), i726-i734. https://doi.org/10.1093/bioinformatics/btaa805
Schmidt M. L., Kuzmanoff K. L., Ling-Indeck L., Pezzuto J. M. (1997). Betulinic acid induces apoptosis in human neuroblastoma cell lines. European Journal of Cancer, 33(12), 2007-2010. https://doi.org/10.1016/S0959-8049(97)00294-3
Seaver B., Smith J. R. (2004). Inhibition of COX isoforms by nutraceuticals. Journal of Herbal Pharmacotherapy, 4(2), 11-18. https://doi.org/10.1080/J157v04n02_02
Serin G., Joseph G., Ghisolfi L., Bauzan M., Erard M., Amalric F., Bouvet P. (1997). Two RNA-binding domains determine the RNA-binding specificity of nucleolin. Journal of Biological Chemistry, 272(20), 13109-13116. https://doi.org/10.1074/jbc.272.20.13109
Shen G., Jeong W. S., Hu R., Kong A. N. T. (2005). Regulation of Nrf2, NF-κB, and AP-1 signaling pathways by chemopreventive agents. Antioxidants & redox signaling, 7(11-12), 1648-1663.https://doi.org/10.1089/ars.2005.7.1648
Sun Y., Xun K., Wang Y., Chen X. (2009). A systematic review of the anticancer properties of berberine, a natural product from Chinese herbs. Anti-Cancer Drugs, 20(9), 757-769. https://doi.org/10.1097/CAD.0b013e328330d95b
Tajrishi M. M., Tuteja R., Tuteja N. (2011). Nucleolin: The most abundant multifunctional phos-phoprotein of nucleolus. Communicative & Integrative Biology, 4(3), 267-275. https://doi.org/10.4161/cib.4.3.14884
Thurnher D., Turhani D., Pelzmann M., Wannemacher B., Knerer B., Formanek M., Wacheck V., Selzer E. (2003). Betulinic acid: a new cytotoxic compound against malignant head and neck cancer cells. Head & Neck: Journal for the Sciences and Specialties of the Head and Neck, 25 (9), 732-740. https://doi.org/10.1002/hed.10231
Trott O., Olson A. J. (2010). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31(2), 455-461. https://doi.org/10.1002/jcc.21334.
Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., Mackerell A. D. (2010). CHARMM general force field: A force field for druglike molecules compatible with the CHARMM all-atom additive biological force fields. Journal of Computational Chemistry, 31(4), 671-690. https://doi.org/10.1002/jcc.21367
Vasaturo M., Cotugno R., Fiengo L., Vinegoni C., Dal Piaz F., De Tommasi N. (2018). The anti-tumor diterpene oridonin is a direct inhibitor of Nucleolin in cancer cells. Scientific reports, 8(1), 1-13. https://doi.org/10.1038/s41598-018-35088-x
Volkamer A., Kuhn D., Grombacher T., Rippmann F., Rarey M. (2012). Combining Global and Local Measures for Structure Based Druggability Predictions. Journal of Chemical Information and Modeling, 52(2), 360-372. https://doi.org/10.1021/ci200454v
Walters, W. P. (2012). Going further than Lipinski’s rule in drug design. Expert opinion on drug discovery, 7(2), 99-107. https://doi.org 10.1517/17460441.2012.648612
Wang J., Wu J., Li X., Liu H., Qin J., Bai Z., Chi B., Chen X. (2018). Identification and validation nucleolin as a target of curcumol in nasopharyngeal carcinoma cells. Journal of Proteomics, 182, 1-11. https://doi.org/10.1016/j.jprot.2018.04.025
Wang N., Tan H. Y., Li L., Yuen M. F., Feng Y. (2015). Berberine and Coptidis Rhizoma as potential anticancer agents: Recent updates and future perspectives. Journal of Ethnopharmacology, 176, 35-48. https://doi.org/10.1016/j.jep.2015.10.028
Watanabe T., Hirano K., Takahashi A., Yamaguchi K., Beppu M., Fujiki H., Suganuma M. (2010). Nucleolin on the cell surface as a new molecular target for gastric cancer treatment. Biological and Pharmaceutical Bulletin, 33(5), 796-803. https://doi.org/10.1248
/bpb.33.796
Wei, W., Rasul, A., Sadiqa, A., Sarfraz, I., Hussain, G., Nageen, B., Liu, X., Watanabe, N., Selamoglu, Z., Ali, M., Li, X., Li, J. (2019). Curcumol: from plant roots to cancer roots. International Journal of Biological Sciences, 15(8), 1600-1609. https://doi.org/10.7150/ijbs.34716.
Wei, Z. L., Juan, W., Tong, D., Juan, L. X., Sa, L. Y., Jie, H. F. M., Xiao, G., Xiang, L. G., Jie, H. M., Xu, C. (2023). Curcumol inhibits breast cancer growth via NCL/ERα36 and the PI3K/AKT pathway. Food & Function, 14 (2), 74-885. https://doi.org/10.1039/d2fo02387c.
Wick W., Grimmel C., Wagenknecht B., Dichgans J., Weller M. (1999). Betulinic acid-induced apoptosis in glioma cells: A sequential requirement for new protein synthesis, formation of reactive oxygen species, and caspase processing. Journal of Pharmacology and Experimental Therapeutics, 289(3), 1306-1312.
Wise J. F., Berkova Z., Mathur R., Zhu H., Braun F. K., Tao R. H., Sabichi A. L., Ao X., Maeng H., Samaniego F. (2013). Nucleolin inhibits Fas ligand binding and suppresses Fas-mediated apoptosis in vivo via a surface nucleolin-Fas complex. Blood, The Journal of the American Society of Hematology, 121(23), 4729-4739. https://doi.org/10.1182/blood-2012-12-471094
Yan P., Sun X. (2018). Triptolide: A new star for treating human malignancies. Journal of Cancer Research and Therapeutics, 14(9), 271-275. https://doi.org/10.4103/0973-1482.235340
Zhou, P., Huang, J., Tian, F. (2012). Specific noncovalent interactions at protein-ligand interface: implications for rational drug design. Current Medicinal Chemistry, 19(2), 226-238. https://doi.org/10.2174/092986712803414150

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-SinDerivadas 4.0.
Derechos de autor 2023 Revista de la Academia Colombiana de Ciencias Exactas, Físicas y Naturales